Quantitative Reconstruction of Multiscale Imaging Data
Because we take the cell dynamics as the necessary level of integration of living organism's multilevel dynamics, we first focused on imaging and image processing for the reconstruction of cellular dynamics from 3D+time imaging of developing organisms with highlighted nucleus and membrane.
We developed original methods that are now integrated in a workflow architecture operating in part on the European computation grid EGI. The goal is to reconstruct as fast and as well as possible a digital specimen from 3D or 3D+time data for further quantitative analysis and multilevel modeling.
The workflow architecture is made for the easy integration of algorithmic modules. As part of its research activity, BioEmergences and its partners in Embryome-DC go on developing new methods to achieve the reconstruction of phenomenological multilevel data. The main methods currently integrated in our workflow in its production mode and available to design specific pipelines include the algorithms below.
Image denoising and enhancement
Image preprocessing is a critical step in order to enhance the information in 3D + time images, often suffering of low contrast and artifacts due to heterogenous distribution of fluorescent proteins. Images are filtered with an edge preserving/enhancing technique, the Geodesic Mean Curvature Flow (GMCF), a PDEs based method able to improve the signal-to-noise ratio, faithfully preserving the position of boundaries that define the shape of the structures.


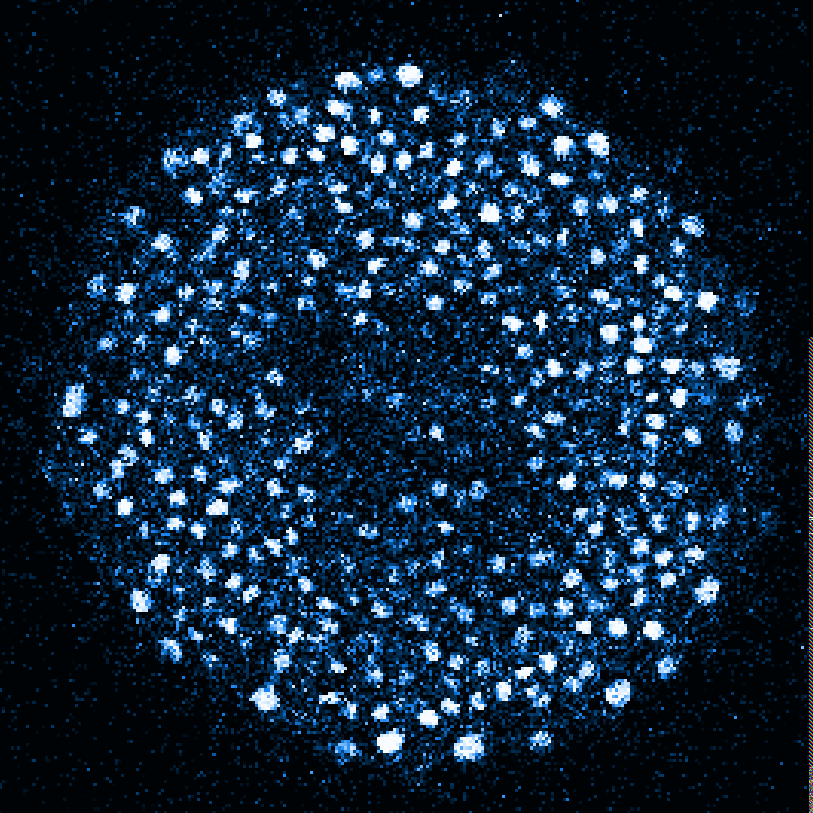
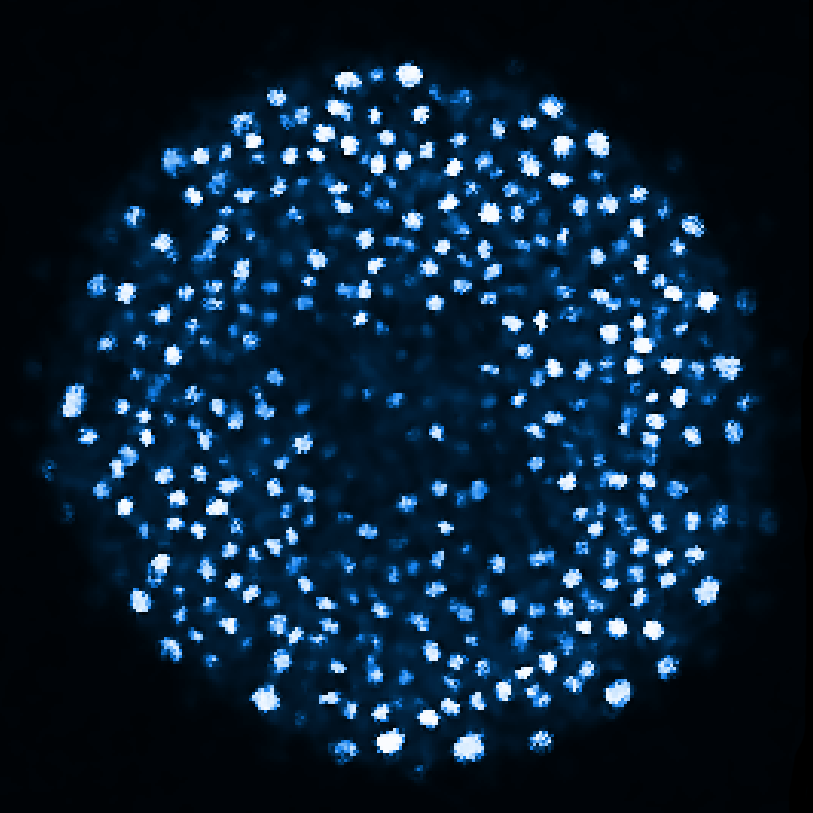
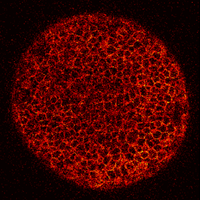
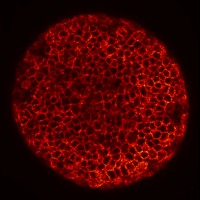
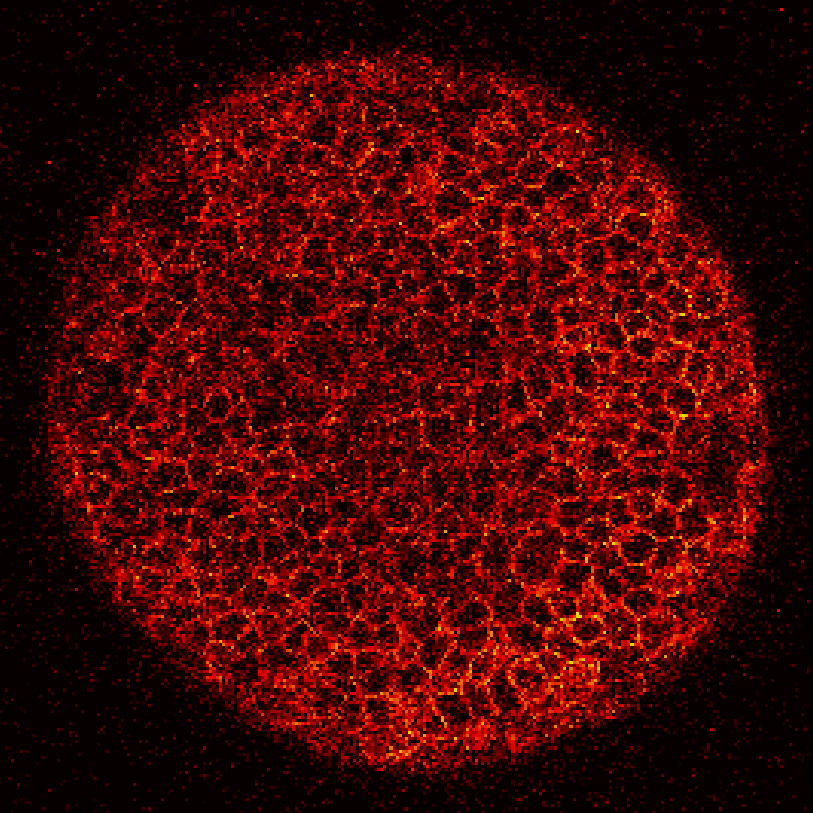
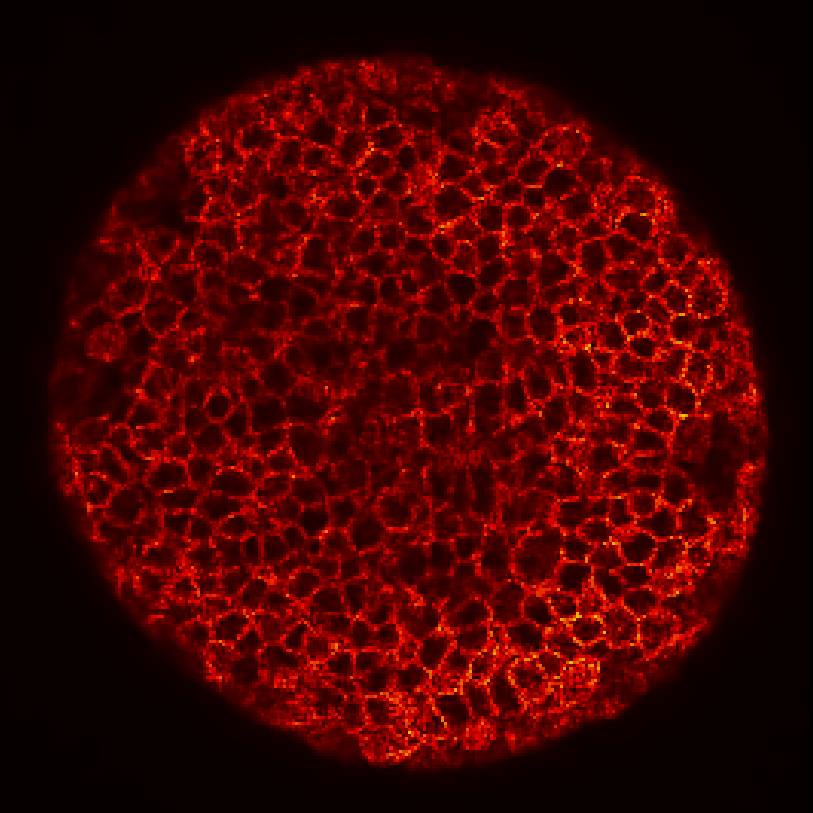
Zebrafish embryo at 5.75 hpf, transverse plane 100 microns deep from the animal pole. Data acquired with a microscope Leica SP5 (980nm and 1030nm 2-photon excitation). Blue, cell nuclei. Red, cell membranes. Blue and red top left, original data. Blue and red top right, filtered data. Blue and red bottom left, ROI of raw images at higher magnification. Blue and red bottom right, ROI of filtered images at higher magnification. The ROI can be selected interactively by moving the mouse over. Click to zoom in/out.
Image registration
State-of-the-art multi-resolution registration methods are provided for realigning the position and the orientation of the specimens along subsequent images by compensating global translation and rotation.
Cell position detection
Cell positions are detected as approximate coordinates of nucleus center, which in turn are located by assuming that objects in the image can be seen as humps of the image intensity. Nuclear centers are identified as local maxima of a simplified version of the images of nuclei, obtained either by a Difference of Gaussian filtering (DoG) or by Flux Based Level set (FBLS).
Cell tracking
The cell tracking relates temporally the identity of the cell in between consecutive time steps. From the initial cell population, these temporal links form a tree whose branches split when a cell divides. The BioEmergences web service currently proposes in its production mode the SimAnn method for reconstructing the spatio-temporal lineage tree. SimAnn is based on the minimization of an elastic-matching heuristic functional of the unknown tracking function by simulated annealing. The latter is designed to assign penalties for large displacements and deformations in cell feature space.
Cell segmentation
Typically, 4-D images acquired from developing organisms provide incomplete information such as cell membranes with missing boundaries and overlapping nuclei. Our strategy for shape segmentation is based on a generalized version of the Subjective Surfaces method (SubSurf) chosen for its ability to complete missing contours.
